Personal motivation: My research interests lie in the physics of living systems. I am fascinated by how effective physics is at reducing the material world to compact equations. When it comes to living systems, their huge complexity seemingly poses a barrier towards finding unifying principles. Yet I believe it is possible to strip down this complexity to its essential elements to understand biological systems with the same depth and rigor physics has brought to our understanding of non-living matter.
Group’s focus: Our aim is to discover general principles that govern living systems using theoretical and computational tools, while remaining close to experimental data, collaborating with experimental groups. We are currently focused on the dynamics of microbial communities, combining theory and data analysis to understand microbiome-environment and host-microbiome relations. Therefore, we also consider the host immune system and how its cells communicate with the microbiome.
Research Focus
Related previous work
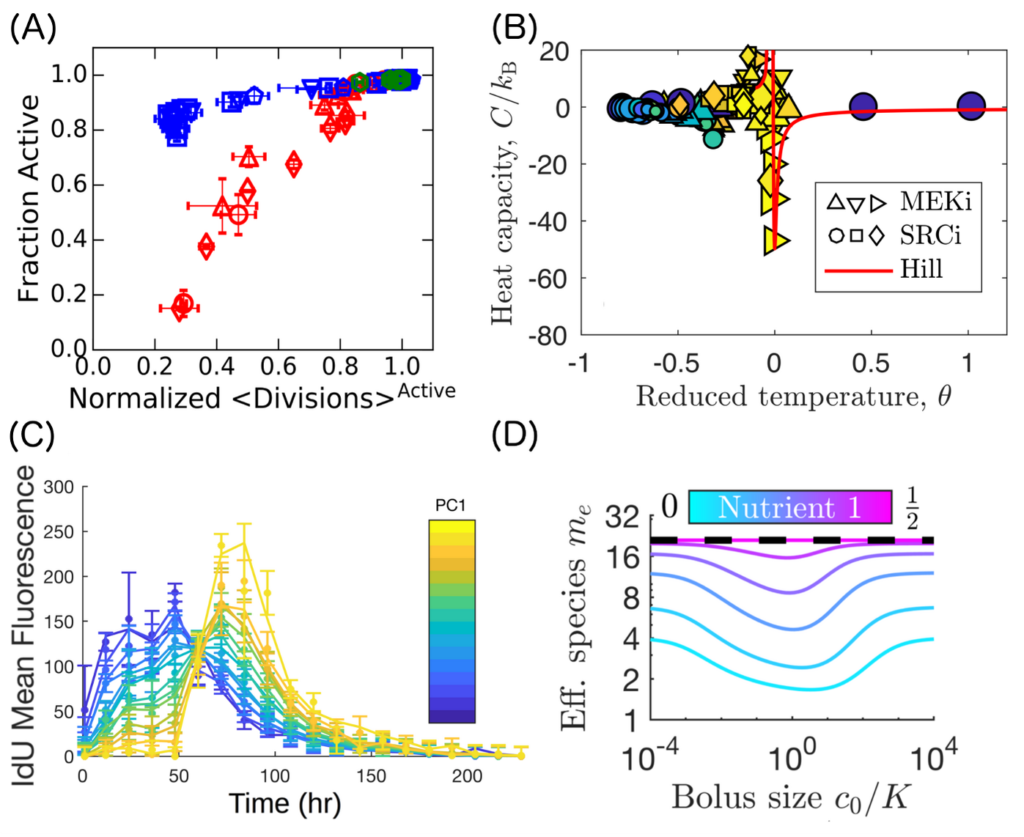
A selection of results from previous research. (A) From Vogel, Erez, Altan-Bonnet (2016): Scaling collapse of experimental data on T-cells. Six different chemotherapy drugs (symbols) in a range of doses collapse to two curves based on their locus of activity in the T-cell signaling network topology: inhibiting before a feedback loop (red) or after it (blue). Inhibiting after feedback can lead to slower cell divisions even for the activated T-cells. (B) From Erez, Byrd, … Mugler (2019): We developed a mapping from a stochastic biochemical feedback model to Ising-like thermodynamic state variables. Applying the mapping to experimental data reveals a diverging heat capacity when cells are manipulated to be near their bifurcation point. Theory (red line) matches experimental data on T-cells (symbols), providing a new method to compute absolute molecule count from flow-cytometry data. (C) From Erez, Mukherjee, Altan-Bonnet (2019): A combination of an IdU pulse-chase experiment (data, symbols) and transfer function theory (fit, lines) along the first principal component (PC1) of phenotype density. Our method directly tracks and estimates differentiation rates of multiple cell populations in the bone marrow, here showing neutrophil maturation from the CD11b-Ly6G- proliferating progenitors to the CD11b+Ly6G+ mature neutrophils. (D) From Erez, Lopez,…,Wingreen (2020): We propose and investigate a model for microbial coexistence in a seasonal ecosystem and show that the effective number of species can depend non-monotonically on the size of the nutrient bolus. Our results imply that the long-sought-after universal relation between species diversity and nutrient amount may not exist but rather depends on ecosystem conditions.